Emergence of Donor-Derived Cell-Free DNA Monitoring For Heart Transplant: Future of Rejection Surveillance
Quick Takes
- Recipient- and donor-derived cell-free DNA (dd-cfDNA) can be detected in the blood of heart transplant recipients, and the proportion of dd-cfDNA increases during acute rejection.
- The increase in dd-cfDNA is seen in both acute cellular rejection (ACR) and antibody-mediated rejection (AMR); ACR and AMR seem to have distinct dd-cfDNA time trajectories, quantitative levels and possibly dd-cfDNA fragment type/length.
- Clinical utility in the use of dd-cfDNA in graft surveillance and potential future uses in altering immunosuppressive therapy based on dd-cfDNA levels requires further study.
Acute rejection, including acute cellular rejection (ACR) and antibody-mediated rejection (AMR), is an anticipated clinical adverse event, especially early (in the first year) after heart transplant (HTx). Since acute rejection can result in graft dysfunction, hemodynamic compromise, chronic graft failure and mortality, accurate and timely diagnosis is essential. The current reference standard for diagnosis of rejection is histological examination of myocardial tissue obtained by endomyocardial biopsy, but this technique is invasive, comes with patient discomfort, potential complications, cost and importantly has its own limitations due to sampling error, interobserver variability, and low sensitivity to detect early rejection.1 As a result, investigations have sought to identify accurate noninvasive methods to detect allograft rejection. Gene expression profiling, breath testing, urine-based assays, and donor-derived cell-free DNA (dd-cfDNA) have been proposed as approaches that may reduce our need for the invasive endomyocardial biopsy.2-4 Here we review the current data on dd-cfDNA in HTx.
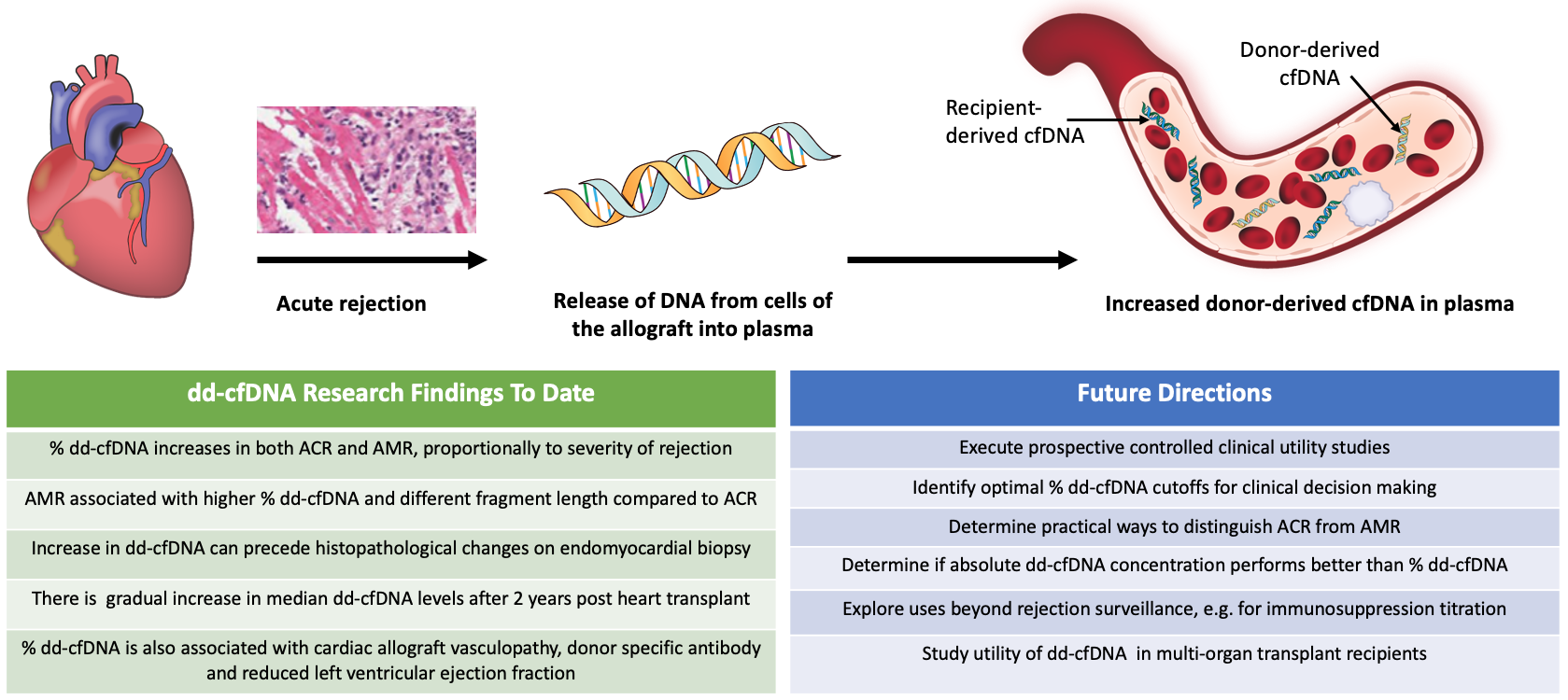
When a cell undergoes necrosis or apoptosis, fragments of double-stranded DNA are released into the tissues and circulation, in the form of cfDNA (DNA not contained within cells). In a solid organ transplant recipient, cfDNA originates from both the recipient's native tissues and from the transplanted donor organ. Therefore, the cfDNA comes from two distinct genomes. Assays that can differentiate cfDNA from two distinct sources – mother and fetus – have been developed for the purposes of prenatal diagnostics. Similarly, assays that can detect donor and recipient cfDNA have been developed and assessed in solid organ transplantation. The main impetus for this was the hypothesis that allograft rejection would result in an increase in circulating dd-cfDNA (Figure 1), and that detection of increased cfDNA would allow for acute rejection surveillance with reduced reliance on endomyocardial biopsy.
Figure 1
There are now several assays that quantify the level of dd-cfDNA after HTx, and the respective studies are consistent in showing that an increase in the dd-cfDNA fraction in plasma correlates with acute graft rejection.5,6 An approach that uses whole genome shotgun sequencing to differentiate between the donor and the recipient genomes has been developed at Stanford University.5 This approach has been tested in a prospective study by De Vlaminck et al. and in a National Institutes of Health (NIH) funded Genomic Research Alliance for Transplantation (GRAfT) consortium.7,8 These studies showed that dd-cfDNA fraction increased in the setting of both ACR and AMR. In AMR, the dd-cfDNA increase started before histopathological changes were detected, and typically reached higher levels than in ACR. The limitation of whole genome sequencing is the need for donor and recipient genotyping and a relatively high cost. An alternative approach used targeted sequencing of highly informative genomic regions, rather than the whole genome, but donor and recipient genotyping was still required.4 Digital droplet polymerase chain reaction (PCR), which is more efficient, also requires donor and/or recipient genotyping.9
Next generation sequencing targeting highly informative single-nucleotide polymorphisms (SNPs) opened a possibility for faster and more efficient testing. A myTAIHEART® assay (myTAIHEART, TAI Diagnostics Inc.) used targeted sequencing of selected 94 SNPs and real-time PCR, but recipient genotyping was still required.10 Lastly, approaches were proposed that would eliminate the need for the genotyping of either the donor or the recipient. AlloSure® assay (CareDx, Inc., South San Francisco, CA) infers the donor and recipient genotype at each of 266 highly polymorphic SNPs and the relative fraction of dd-cfDNA is calculated.11 Data from a prospective registry of this test showed that median dd-cfDNA fraction was 0.07% in samples without rejection and 0.17% in samples with acute rejection.11 This assay is Clinical Laboratory Improvement Amendments (CLIA) validated and commercially available for adult and pediatric HTx populations past 14 days post-transplant. The Prospera™ Heart assay (Natera Inc., Austin, TX) uses 13,000 SNPs and massively multiplexed PCR technology to detect dd-cfDNA; dd-cfDNA fraction of 0.15% was reported to discriminate acute rejection from no rejection.12 This CLIA-validated assay is commercially available and does not require centrifuging of the blood sample or shipment on dry ice.12 Another assay that also uses next generation sequencing and PCR is the Viracor TRAC® assay (Eurofins Scientific, Lee's Summit, MO).13
While there are differences in the technical approaches to detection of dd-cfDNA, the data are overall consistent in that acute graft rejection is associated with rise in dd-cfDNA. The studies show that dd-cfDNA fraction increases in both ACR and AMR, and that the fraction increases proportionally depending on the severity of rejection.7,8,10 The reported sensitivity and specificity of the tests varies by the selected cutoff level and is also influenced by the inherent limitation of endomyocardial biopsy findings. When compared to endomyocardial biopsy surveillance, increase in dd-cfDNA levels occurred 0.5 months before ACR and up to 3.2 months before AMR was detected by biopsy.8 AMR also appears to lead to quantitatively higher %dd-cfDNA levels compared to ACR, and the fragment length of dd-cfDNA was shorter in AMR compared to ACR or no rejection controls.8 Further, %dd-cfDNA was associated with additional clinical variables, including presence of donor specific antibodies, left ventricular ejection fraction, and the presence of cardiac allograft vasculopathy. There also appears to be an overall gradual increase of dd-cfDNA past 2 years after transplant.8
As use of dd-cfDNA across HTx programs increases, there are important questions that still need to be addressed.14-16 While association of %dd-cfDNA with rejection has been established in numerous observational studies, prospective controlled clinical utility studies are lacking. As a result, clinical adoption of dd-cfDNA as a test to reduce the number of endomyocardial biopsies varies widely across centers. Planned controlled prospective studies may provide further guidance on how to best integrate this test to surveillance protocols, and whether additional aspects of the test (e.g., absolute dd-cfDNA concentration vs. %dd-cfDNA) provide useful information. While dd-cfDNA could also assist clinicians in decisions on immunosuppression titration, this too needs to be evaluated prospectively. Whether and how %dd-cfDNA could be used in multi-organ transplantation is also unknown.
In summary, dd-cfDNA has emerged as a promising non-invasive test that is changing our approach to rejection surveillance in HTx. Future studies will likely provide important data on further applications of this test in the care of HTx recipients.
References
- Shah KB, Flattery MP, Smallfield MC, et al. Surveillance endomyocardial biopsy in the modern era produces low diagnostic yield for cardiac allograft rejection. Transplantation 2015;99:e75-80.
- Hollander Z, Mohammadi TH, Assadian S, et al. Cost‐effectiveness of a blood‐based biomarker compared to endomyocardial biopsy for the diagnosis of acute allograft rejection. J Heart Lung Transplant 2016;35:S53.
- Moayedi Y, Foroutan F, Miller RJH, et al. Risk evaluation using gene expression screening to monitor for acute cellular rejection in heart transplant recipients. J Heart Lung Transplant 2019;38:51‐58.
- Hidestrand M, Tomita‐Mitchell A, Hidestrand PM, et al. Highly sensitive noninvasive cardiac transplant rejection monitoring using targeted quantification of donor‐specific cell‐free deoxyribonucleic acid. J Am Coll Cardiol 2014;63:1224‐26.
- Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A 2011;108:6229-34.
- Beck J, Oellerich M, Schulz U, et al. Donor‐derived cell‐free DNA is a novel universal biomarker for allograft rejection in solid organ transplantation. Transplant Proc 2015;47:2400‐03.
- De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77.
- Agbor-Enoh S, Shah P, Tunc I, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation 2021;143:1184-97.
- Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem 2013;59:1732-41.
- North PE, Ziegler E, Mahnke DK, et al. Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: validation of a rapid and highly sensitive clinical test for stratification of rejection probability. PLoS One 2020;15:e0227385.
- Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant 2019;19:2889-99.
- Natera Announces the Validation and Launch of the Prospera™ Heart Transplant Assessment Test (Natera.com). 2021. Available at: https://www.natera.com/company/news/natera-announces-the-validation-and-launch-of-the-prospera-heart-transplant-assessment-test/. Accessed 01/18/2022.
- Viracor TRAC® Heart dd-cfDNA (eurofins-viracor.com). 2022. Available at: https://www.eurofins-viracor.com/clinical/test-menu/30877-viracor-trac-heart-dd-cfdna/. Accessed 01/30/2022.
- Zhang H, Lui L, Zheng C, et al. The role of donor-derived cell-free DNA in the identification of injury in kidney allografts with antibody-mediated rejection or de novo DSA. Transplantation 2018;102:S5-6.
- Holzhauser L, Fujino T, Alenghat F, et al. Donor-derived cell-free DNA is associated with cardiac allograft vasculopathy. J Heart Lung Transpl 2020;39:S63.
- Crespo-Leiro MG, Hiller D, Woodward RN, et al. Analysis of donor-derived cell-free DNA with 3-year outcomes in heart transplant recipients. J Heart Lung Transplant 2017;36:S69-70.
Clinical Topics: Arrhythmias and Clinical EP, Cardiac Surgery, Cardiovascular Care Team, Invasive Cardiovascular Angiography and Intervention, Genetic Arrhythmic Conditions, Cardiac Surgery and Arrhythmias, Cardiac Surgery and Heart Failure, Novel Agents, Heart Transplant, Heart Failure and Cardiomyopathies
Keywords: Stroke Volume, Prospective Studies, Graft Rejection, Polymorphism, Single Nucleotide, Genotype, High-Throughput Nucleotide Sequencing, Observer Variation, Real-Time Polymerase Chain Reaction, Selection Bias, Ventricular Function, Left, DNA, Cell-Free Nucleic Acids, Allografts, Heart Transplantation, Biopsy, Reference Standards, Apoptosis, National Institutes of Health (U.S.), Gene Expression Profiling, Whole Genome Sequencing, Nucleotides, Genomics, Necrosis, Registries
< Back to Listings